RNA Peach and Mango: Orthogonal two-color fluorogenic aptamers distinguish nearly identical ligands.
Two-channel fluorogenic RNA aptamer-based imaging is currently challenging. While we have previously characterized the Mango series of aptamers that bind tightly and specifically to the green fluorophore TO1-Biotin, the next aim was to identify an effective fluorogenic aptamer partner for two-color imaging. A competitive in vitro selection for TO3-Biotin binding aptamers was performed resulting in the Peach I and II aptamers. Remarkably, given that the TO1-Biotin and TO3-Biotin heterocycles differ by only two bridging carbons, these new aptamers exhibit a marked preference for TO3-Biotin binding relative to the iM3 and Mango III A10U aptamers, which preferentially bind TO1-Biotin. Peach I, like Mango I and II, appears to contain a quadruplex core isolated by a GAA^A type tetraloop-like adaptor from its closing stem. Thermal melts of the Peach aptamers reveal that TO3-Biotin binding is cooperative, while TO1-Biotin binding is not, suggesting a unique and currently uncharacterized mode of ligand differentiation. Using only fluorescent measurements, the concentrations of Peach and Mango aptamers could be reliably determined in vitro. The utility of this orthogonal pair provides a possible route to in vivo two-color RNA imaging.
Read MoreRNA molecules play vital roles in many cellular processes. Visualising their dynamics in live cells at single-molecule resolution is essential to elucidate their role in RNA metabolism. RNA aptamers, such as Spinach and Mango, have recently emerged as a powerful background-free technology for live-cell RNA imaging due to their fluorogenic properties upon ligand binding. Here, we report a novel array of Mango II aptamers for RNA imaging in live and fixed cells with high contrast and single-molecule sensitivity.
Read MoreSeveral turn-on RNA aptamers that activate small-molecule fluorophores have been selected in vitro. Among these, the ~30 nucleotide Mango-III is notable because it binds the thiazole orange derivative TO1-Biotin with high affinity and fluoresces brightly (quantum yield 0.55). Uniquely among related aptamers, Mango-III exhibits biphasic thermal melting, characteristic of molecules with tertiary structure. We report crystal structures of TO1-Biotin complexes of Mango-III, a structure-guided mutant Mango-III(A10U), and a functionally reselected mutant iMango-III.
Read MoreHere we present a sensitive, rapid, and discriminating post-gel staining method to image RNAs tagged with RNA Mango aptamers I, II, III, or IV, using either native or denaturing polyacrylamide gel electrophoresis (PAGE) gels. After running standard PAGE gels, Mango-tagged RNA can be easily stained with TO1-Biotin and then analyzed using commonly available fluorescence readers.
Read MoreThere is a pressing need for nucleic acid-based assays that are capable of rapidly and reliably detecting pathogenic organisms. Many of the techniques available for the detection of pathogenic RNA possess one or more limiting factors that make the detection of low-copy RNA challenging. While RT-PCR is the most commonly used method for detecting pathogen-related RNA, it requires expensive thermocycling equipment and is comparatively slow. Isothermal methods promise procedural simplicity, but have traditionally suffered from amplification artifacts that tend to preclude easy identification of target nucleic acids. Recently, the isothermal SHERLOCK system overcame this problem by using CRISPR to distinguish amplified target sequences from artifactual background signal. However, this system comes at the cost of introducing considerable enzymatic complexity and corresponding increase in total assay time. Therefore, simpler and less expensive strategies are highly desirable. Here, we demonstrate that by nesting NASBA primers and modifying the NASBA inner primers to encode an RNA Mango aptamer sequence we can dramatically increase the sensitivity of NASBA to 1.5 RNA molecules per microliter. As this isothermal nucleic acid detection scheme directly produces a fluorescent reporter, real-time detection is intrinsic to the assay. Nested Mango NASBA is highly specific and, in contrast to existing RNA detection systems, offers a cheap, simple, and specific way to rapidly detect single molecule amounts of pathogenic RNA.
Read MoreSeveral RNA aptamers that bind small molecules and enhance their fluorescence have been successfully used to tag and track RNAs in vivo, but these genetically encodable tags have not yet achieved single-fluorophore resolution. Recently, Mango-II, an RNA that binds TO1-Biotin with ∼1 nM affinity and enhances its fluorescence by >1500-fold, was isolated by fluorescence selection from the pool that yielded the original RNA Mango. We determined the crystal structures of Mango-II in complex with two fluorophores, TO1-Biotin and TO3-Biotin, and found that despite their high affinity, the ligands adopt multiple distinct conformations, indicative of a binding pocket with modest stereoselectivity. Mutational analysis of the binding site led to Mango-II(A22U), which retains high affinity for TO1-Biotin but now discriminates >5-fold against TO3-biotin. Moreover, fluorescence enhancement of TO1-Biotin increases by 18%, while that of TO3-Biotin decreases by 25%. Crystallographic, spectroscopic, and analogue studies show that the A22U mutation improves conformational homogeneity and shape complementarity of the fluorophore–RNA interface. Our work demonstrates that even after extensive functional selection, aptamer RNAs can be further improved through structure-guided engineering.
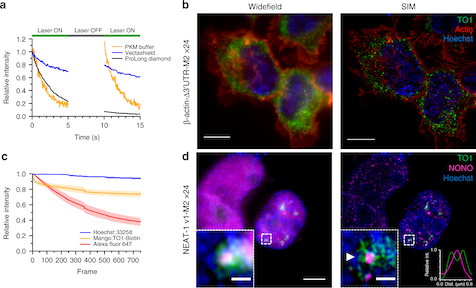
Read MoreDespite having many key roles in cellular biology, directly imaging biologically important RNAs has been hindered by a lack of fluorescent tools equivalent to the fluorescent proteins available to study cellular proteins. Ideal RNA labelling systems must preserve biological function, have photophysical properties similar to existing fluorescent proteins, and be compatible with established live and fixed cell protein labelling strategies. Here, we report a microfluidics-based selection of three new high-affinity RNA Mango fluorogenic aptamers. Two of these are as bright or brighter than enhanced GFP when bound to TO1-Biotin. Furthermore, we show that the new Mangos can accurately image the subcellular localization of three small non-coding RNAs (5S, U6, and a box C/D scaRNA) in fixed and live mammalian cells. These new aptamers have many potential applications to study RNA function and dynamics both in vitro and in mammalian cells.
Read MoreThe characterization of RNA-protein complexes (RNPs) is a difficult but increasingly important problem in modern biology. By combining the compact RNA Mango aptamer with a fluorogenic thiazole orange desthiobiotin (TO1-Dtb or TO3-Dtb) ligand, we have created an RNA tagging system that simplifies the purification and subsequent characterization of endogenous RNPs. Mango-tagged RNP complexes can be immobilized on a streptavidin solid support and recovered in their native state by the addition of free biotin. Furthermore, Mango-based RNP purification can be adapted to different scales of RNP isolation ranging from pull-down assays to the isolation of large amounts of biochemically defined cellular RNPs. We have incorporated the Mango aptamer into the S. cerevisiae U1 small nuclear RNA (snRNA), shown that the Mango-snRNA is functional in cells, and have used the aptamer to pull-down a U1 snRNA associated protein. To demonstrate large scale isolation of RNPs, we purified and characterized bacterial RNA polymerase holoenzyme (HE) in complex with a Mango-containing 6S RNA. We were able to use the combination of a red shifted TO3-Dtb ligand and eGFP tagged HE to follow the binding and release of the 6S RNA by two color native gel analysis as well as by single-molecule fluorescence cross-correlation spectroscopy. Together these experiments demonstrate how the Mango aptamer in conjunction with simple derivatives of its flurophore ligands enables the purification and characterization of endogenous cellular RNPs in vitro.
Read MoreGenetically encoded fluorescent protein tags have revolutionized proteome studies, whereas the lack of intrinsically fluorescent RNAs has hindered transcriptome exploration. Among several RNA–fluorophore complexes that potentially address this problem, RNA Mango has an exceptionally high affinity for its thiazole orange (TO)-derived fluorophore, TO1–Biotin (Kd ∼3 nM), and, in complex with related ligands, it is one of the most redshifted fluorescent macromolecular tags known. To elucidate how this small aptamer exhibits such properties, which make it well suited for studying low-copy cellular RNAs, we determined its 1.7-Å-resolution co-crystal structure. Unexpectedly, the entire ligand, including TO, biotin and the linker connecting them, abuts one of the near-planar faces of the three-tiered G-quadruplex. The two heterocycles of TO are held in place by two loop adenines and form a 45° angle with respect to each other. Minimizing this angle would increase quantum yield and further improve this tool for in vivo RNA visualization..
Read MoreThe effective tracking and purification of biological RNAs and RNA protein complexes is currently challenging. One promising strategy to simultaneously address both of these problems is to develop high-affinity RNA aptamers against taggable small molecule fluorophores. RNA Mango is a 39-nucleotide, parallel-stranded G-quadruplex RNA aptamer motif that binds with nanomolar affinity to a set of thiazole orange (TO1) derivatives while simultaneously inducing a 103-fold increase in fluorescence. We find that RNA Mango has a large increase in its thermal stability upon the addition of its TO1-Biotin ligand. Consistent with this thermal stabilization, RNA Mango can effectively discriminate TO1-Biotin from a broad range of small molecule fluorophores. In contrast, RNA Spinach, which is known to have a substantially more rigid G-quadruplex structure, was found to bind to this set of fluorophores, often with higher affinity than to its native ligand, 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI), and did not exhibit thermal stabilization in the presence of the TO1-Biotin fluorophore. Our data suggest that RNA Mango is likely to use a concerted ligand-binding mechanism that allows it to simultaneously bind and recognize its TO1-Biotin ligand, whereas RNA Spinach appears to lack such a mechanism. The high binding affinity and fluorescent efficiency of RNA Mango provides a compelling alternative to RNA Spinach as an RNA reporter system and paves the way for the future development of small fluorophore RNA reporter systems.
Read MoreBecause RNA lacks strong intrinsic fluorescence, it has proven challenging to track RNA molecules in real time. To address this problem and to allow the purification of fluorescently tagged RNA complexes, we have selected a high affinity RNA aptamer called RNA Mango. This aptamer binds a series of thiazole orange (fluorophore) derivatives with nanomolar affinity, while increasing fluorophore fluorescence by up to 1,100-fold. Visualization of RNA Mango by single-molecule fluorescence microscopy, together with injection and imaging of RNA Mango/fluorophore complex in C. elegans gonads demonstrates the potential for live-cell RNA imaging with this system. By inserting RNA Mango into a stem loop of the bacterial 6S RNA and biotinylating the fluorophore, we demonstrate that the aptamer can be used to simultaneously fluorescently label and purify biologically important RNAs. The high affinity and fluorescent properties of RNA Mango are therefore expected to simplify the study of RNA complexes.
Read More