RNA origami is a framework for the modular design of nanoscaffolds that can be folded from a single strand of RNA and used to organize molecular components with nanoscale precision. The design of genetically expressible RNA origami, which must fold cotranscriptionally, requires modelling and design tools that simultaneously consider thermodynamics, the folding pathway, sequence constraints and pseudoknot optimization. Here, we describe RNA Origami Automated Design software (ROAD), which builds origami models from a library of structural modules, identifies potential folding barriers and designs optimized sequences. Using ROAD, we extend the scale and functional diversity of RNA scaffolds, creating 32 designs of up to 2,360 nucleotides, five that scaffold two proteins, and seven that scaffold two small molecules at precise distances. Micrographic and chromatographic comparisons of optimized and non-optimized structures validate that our principles for strand routing and sequence design substantially improve yield. By providing efficient design of RNA origami, ROAD may simplify the construction of custom RNA scaffolds for nanomedicine and synthetic biology.
Read MoreFluorescent RNA aptamers are useful as markers for tracking RNA molecules inside cells and for creating biosensor devices. Förster resonance energy transfer (FRET) based on fluorescent proteins has been used to detect conformational changes, however, such FRET devices have not yet been produced using fluorescent RNA aptamers. Here we develop an RNA aptamer-based FRET (apta-FRET) system using single-stranded RNA origami scaffolds. To obtain FRET, the fluorescent aptamers Spinach and Mango are placed in close proximity on the RNA scaffolds and a new fluorophore is synthesized to increase spectral overlap. RNA devices that respond to conformational changes are developed, and finally, apta-FRET constructs are expressed in E. coli where FRET is observed, demonstrating that the apta-FRET system is genetically encodable and that the RNA nanostructures fold correctly in bacteria. We anticipate that the RNA apta-FRET system could have applications as ratiometric sensors for real-time studies in cell and synthetic biology.
Read MoreRecent advances in fluorogen-binding “light-up” RNA aptamers have enabled protein-free detection of RNA in cells. Detailed biophysical characterization of folding of G-Quadruplex (GQ)-based light-up aptamers such as Spinach, Mango and Corn is still lacking despite the potential implications on their folding and function. In this work we employ single-molecule fluorescence-force spectroscopy to examine mechanical responses of Spinach2, iMangoIII and MangoIV. Spinach2 unfolds in four discrete steps as force is increased to 7 pN and refolds in reciprocal steps upon force relaxation. In contrast, GQ-core unfolding in iMangoIII and MangoIV occurs in one discrete step at forces >10 pN and refolding occurred at lower forces showing hysteresis. Co-transcriptional folding using superhelicases shows reduced misfolding propensity and allowed a folding pathway different from refolding. Under physiologically relevant pico-Newton levels of force, these aptamers may unfold in vivo and subsequently misfold. Understanding of the dynamics of RNA aptamers will aid engineering of improved fluorogenic modules for cellular applications.
Read MoreBacteriophage Q protein engages σ-dependent paused RNA polymerase (RNAP) by binding to a DNA site embedded in late gene promoter and renders RNAP resistant to termination signals. Here, we report a single-particle cryo-electron microscopy (cryo-EM) structure of an intact Q-engaged arrested complex. The structure reveals key interactions responsible for σ-dependent pause, Q engagement, and Q-mediated transcription antitermination. The structure shows that two Q protomers (QI and QII) bind to a direct-repeat DNA site and contact distinct elements of the RNA exit channel. Notably, QI forms a narrow ring inside the RNA exit channel and renders RNAP resistant to termination signals by prohibiting RNA hairpin formation in the RNA exit channel. Because the RNA exit channel is conserved among all multisubunit RNAPs, it is likely to serve as an important contact site for regulators that modify the elongation properties of RNAP in other organisms, as well.
Read MoreThe MerR-family transcription factors (TFs) are a large group of bacterial proteins responding to cellular metal ions and multiple antibiotics by binding within central RNA polymerase-binding regions of a promoter. While most TFs alter transcription through protein–protein interactions, MerR TFs are capable of reshaping promoter DNA. To address the question of which mechanism prevails, we determined two cryo-EM structures of transcription activation complexes (TAC) comprising Escherichia coli CueR (a prototype MerR TF), RNAP holoenzyme and promoter DNA. The structures reveal that this TF promotes productive promoter–polymerase association without canonical protein–protein contacts seen between other activator proteins and RNAP. Instead, CueR realigns the key promoter elements in the transcription activation complex by clamp-like protein–DNA interactions: these induce four distinct kinks that ultimately position the −10 element for formation of the transcription bubble. These structural and biochemical results provide strong support for the DNA distortion paradigm of allosteric transcriptional control by MerR TFs.
Read MoreMicroRNAs (miRNAs) play essential roles in regulating gene expression and cell fate. However, it remains a great challenge to image miRNAs with high accuracy in living cells. Here, we report a novel genetically encoded dual-color light-up RNA sensor for ratiometric imaging of miRNAs using Mango as an internal reference and SRB2 as the sensor module. This genetically encoded sensor is designed by expressing a splittable fusion of the internal reference and sensor module under a single promoter. This design strategy allows synchronous expression of the two modules with negligible interference. Live cell imaging studies reveal that the genetically encoded ratiometric RNA sensor responds specifically to mir-224. Moreover, the sensor-to-Mango fluorescence ratios are linearly correlated with the concentrations of mir-224, confirming their capability of determining mir-224 concentrations in living cells. Our genetically encoded light-up RNA sensor also enables ratiometric imaging of mir-224 in different cell lines. This strategy could provide a versatile approach for ratiometric imaging of intracellular RNAs, affording powerful tools for interrogating RNA functions and abundance in living cells.
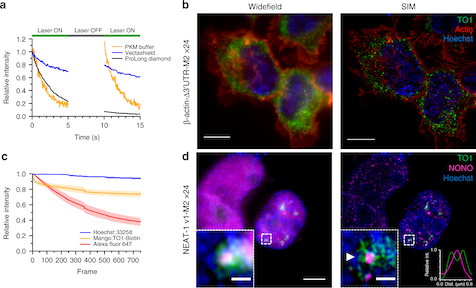
Read MoreDevelopment of biocompatible tools for intracellular imaging of RNA expression remains a central challenge. Herein, we report the use of heterochiral strand-displacement to sequence-specifically interface endogenous d-miRNAs with an l-RNA version of the fluorogenic aptamer Mango III, thereby generating a novel class of biocompatible miRNA sensors. Fluorescence activation of the sensor is achieved through the displacement of an achiral blocking strand from the l-Mango aptamer by the d-RNA target. In contrast to d-Mango, we show that the l-Mango sensor retains full functionality in serum, enabling a light-up fluorescence response to the target. Importantly, we employ a self-delivering version of the l-Mango sensor to image the expression of microRNA-155 in living cells, representing the first time l-oligonucleotides have been interfaced with a living system. Overall, this work provides a new paradigm for the development of biocompatible hybridization-based sensors for live-cell imaging of RNAs and greatly expands the utility of fluorogenic aptamers for cellular applications.
Read More